15 Create
In this guide, we explore how to generate a GeoLocator Data Package from a GeoPressureTemplate folder where you’ve already performed the analysis of the tracks following the GeoPressureTemplate workflow.
To be able to follow this guide, you need to have a GeoPressureTemplate project containing:
- An up-to-date
DESCRIPTIONfile, includingversion,contributors,licencesas described in the R packages book. - One interim
.RDatafile per tag, with all outputs you would like to publish, generated with the Geopressuretemplate workflow. - The
tag.csvandobservations.csvfiles in thedata/folder.
If you’ve not yet performed the GeoPressureR analysis, it is still possible to create a geolocator data package with only the core ressources.
The easiest way to create a Zendo record and ensure that the data is correctly sync is using the Zenodo API and the Zen4R package.
For this, you first need to create a token and save it to your keyring with:
keyring::key_set_with_value("ZENODO_PAT", password = "{your_zenodo_token}")This allows us to create a ZenodoManager object which becomes useful later.
zenodo <- ZenodoManager$new(token = keyring::key_get(service = "ZENODO_PAT"))✔ Successfully connected to Zenodo with user token15.1 Setup of the example
For this example, we won’t be using the Swainson’s Warbler nor Great Reed Warbler as we want a full project with multiple tags. Instead, we download the GeoPressureTemplate of the Woodland Kingfisher geolocator project. Our goal will essentially be to reproduce the associated GeoLocator Data Package uploaded on Zenodo already.
We can download this GeoPressureTemplate from Github in a temporary folder using the following code:
Code
repo <- "Rafnuss/WoodlandKingfisher"
# Create temporary file for the ZIP
temp_zip <- tempfile(fileext = ".zip")
# Download the ZIP file and unzip the repository
download.file(
glue::glue("https://github.com/{repo}/archive/refs/heads/main.zip"),
temp_zip,
mode = "wb"
)
temp_dir <- tempfile()
unzip(temp_zip, exdir = temp_dir)
# return the extraction directory
directory <- file.path(temp_dir, list.files(temp_dir))15.2 Initiate datapackage with metadata
15.2.1 Initiate datapackage using the DESCRIPTION file
We can generate a GeoLocator Data Package pkg from an existing GeoPressureTemplate folder with create_gldp_geopressuretemplate(). This function will read the DESCRIPTION file and create a frictionless package with the metadata you’ve already entered.
pkg <- create_gldp_geopressuretemplate(directory = directory)15.2.2 Create record on Zenodo
The easiest way to update the metadata is to create an empty Zenodo record and fill in all metadata directly on Zenodo. This way, you can benefit from the Zenodo interface to enter all metadata and share it with co-authors for review before creating the package.
z <- zenodo$depositRecord(
gldp_to_zenodo(pkg),
reserveDOI = TRUE,
publish = FALSE
)
browseURL(z$links$self_html)15.2.3 Update metadata on Zenodo
Login to Zenodo and fill in all metadata you want to add to your GeoLocator Data Package. Read the datapackage specification to learn about all recommended metadata that can be added
Note that a datapackage contributors corresponds to creators on Zenodo and not the contributors.
To be able to save the draft of your record, you’ll need to upload some file, even a dummy text file will do. You can always delete it later and upload with the correct ones.
Sharing the draft with co-authors is a great way to ensure that all metadata are correct before publication.
15.2.4 Create datapackage from Zenodo record
Once you’re happy with the metadata entered on Zenodo, you create a new GeoLocator Data Package from the Zenodo record. For this, you need to retrieve the concept DOI of your Zenodo record. The concept DOI is the one that doesn’t change with new versions. The DOI displayed on Zenodo is actually the DOI of the first version, but you can retrieve the concept DOI by substracting 1 to your ID number.
z <- zenodo$getDepositionByConceptDOI("10.5281/zenodo.{ZENODO_ID - 1}")
pkg <- zenodo_to_gldp(z)15.3 Add the data
Now that we have the metadata set up, we can add the data to the package. add_gldp_geopressuretemplate() is a key function that read the geopressuretemplate data available and automatically add the most up-to-date available data to the datapackage.
pkg <- add_gldp_geopressuretemplate(pkg, directory = directory)✔ Reading tags from '/var/folders/cm/dh8lmmxn3b5bcb2_gbz16ycm0000gn/T//Rtmpl5uh1z/file575260807362/WoodlandKingfisher-main/data/tags.csv'.✔ Reading observations from '/var/folders/cm/dh8lmmxn3b5bcb2_gbz16ycm0000gn/T//Rtmpl5uh1z/file575260807362/WoodlandKingfisher-main/data/observations.csv'.15.4 Data to include
By default, add_gldp_geopressuretemplate() adds first the tag data from the data/interim Rdata file. For all tag_id that don’t have a interim file, it read the data directly from the data/raw-data folder, using config.yml information. Using the from argument, you can specify to only read from one of these two locations. It’s possible to exclude specific tag_id by adding a _ to the interim file name or raw-data folder (e.g., _TAG1234.RData or _TAG1234/).
15.4.2 Inspect the created package
Before uploading the data, you should also inspect the created package to ensure that everything is correct.
Check the summary of the package, in particular the number of tags in each resources.
print(pkg)── A GeoLocator Data Package (vmain) • title: "Migration route and timing of South African Woodland Kingfisher"• contributors: Raphaël Nussbaumer ('rafnuss@gmail.com') (Researcher, ProjectLeader) -
<https://orcid.org/0000-0002-8185-1020> Yann Rime ('yann.rime@vogelwarte.ch') (Researcher, Contributor) -
<https://orcid.org/0009-0005-7264-6753>• embargo: 1970-01-01• licenses: Creative Commons Attribution 4.0 (CC-BY-4.0) -
<https://creativecommons.org/licenses/by/4.0/>• created: 2025-12-11 07:59:11• bibliographicCitation: "Nussbaumer R, Rime Y (2025). \"Migration route and
timing of South African Woodland Kingfisher.\""• spatial: Polygon and c(17.375, 32.875, 32.875, 17.375, 17.375, -24.375,
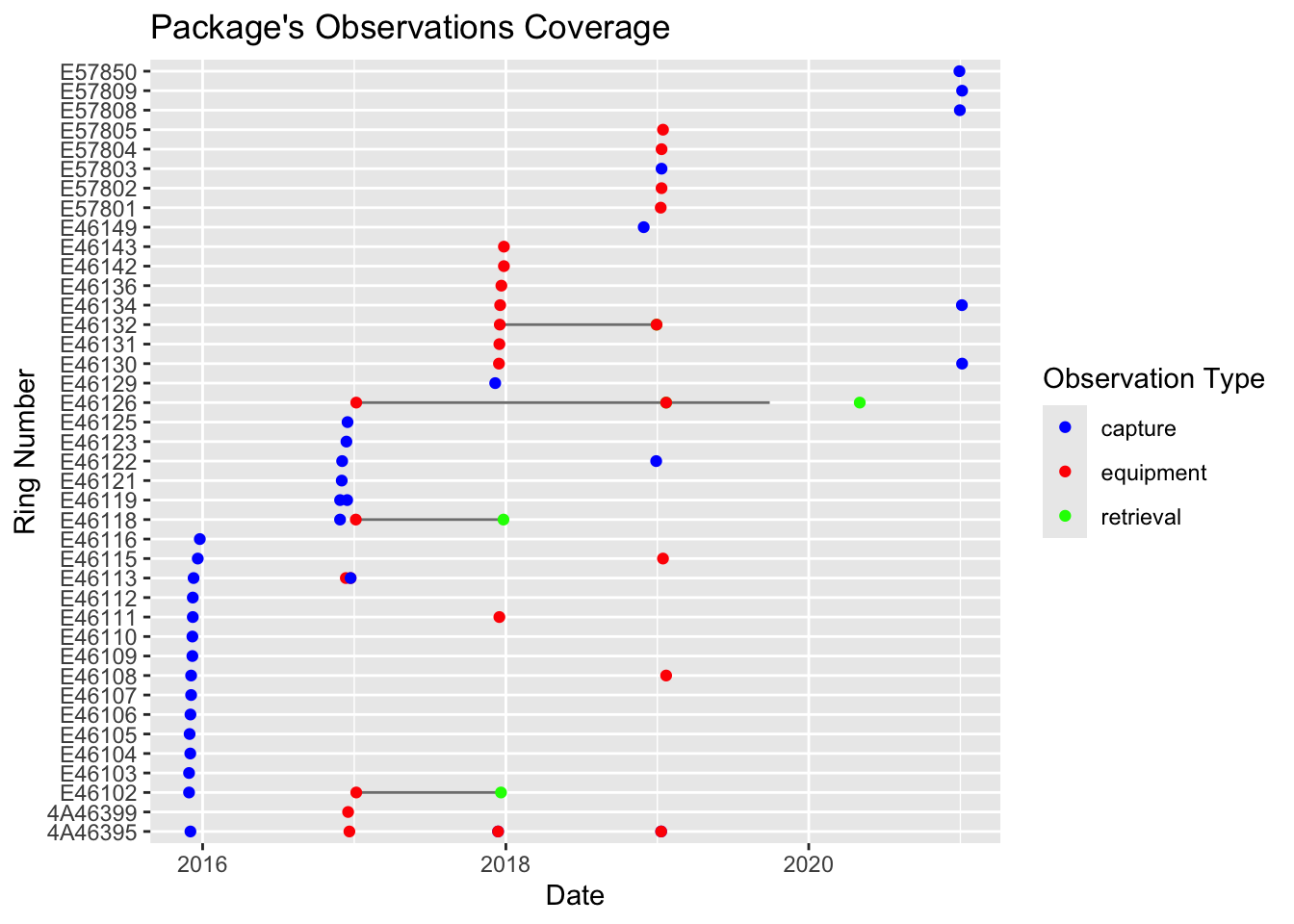
-24.375, 12.875, 12.875, -24.375)• temporal: "2017-01-10" to "2019-09-28"• taxonomic: "Halcyon senegalensis"• numberTags: tags: 25 measurements: 5 light: 5 pressure: 5 activity: 5 temperature_external: 5 magnetic: 5 paths: 5 pressurepaths: 5── 8 resources • twilights (n=3,262)• staps (n=223)• paths (n=22,523)• edges (n=22,018)• pressurepaths (n=224,828)• tags (n=25)• observations (n=64)• measurements (n=1,924,975)Use `unclass()` to print the Geolocator Data Package as a list.You can visualize the coverage of the package by ploting the Data Package. Check for the correct number of tags, as well as equipment and retrieval dates.
Finally, we check that the package is compliant with GeoLocator Data Package standards.
validate_gldp(pkg)
##
## ── Check GeoLocator DataPackage profile ──
##
## ✔ title is valid.
## ✔ contributors is valid.
## ✔ contributors[[1]] is valid.
## ✔ contributors[[2]] is valid.
## ✔ embargo is valid.
## ✔ licenses is valid.
## ✔ licenses[[1]] is valid.
## ✔ created is valid.
## ✔ numberTags is valid.
## ✔ numberTags$tags is valid.
## ✔ numberTags$measurements is valid.
## ✔ numberTags$light is valid.
## ✔ numberTags$pressure is valid.
## ✔ numberTags$activity is valid.
## ✔ numberTags$temperature_external is valid.
## ✔ numberTags$temperature_internal is valid.
## ✔ numberTags$magnetic is valid.
## ✔ numberTags$wet_count is valid.
## ✔ numberTags$conductivity is valid.
## ✔ numberTags$paths is valid.
## ✔ numberTags$pressurepaths is valid.
## ✔ bibliographicCitation is valid.
## ✔ $schema is valid.
## ✔ resources is valid.
## ✔ resources[[1]] is valid.
## ✔ resources[[2]] is valid.
## ✔ resources[[3]] is valid.
## ✔ resources[[4]] is valid.
## ✔ resources[[5]] is valid.
## ✔ resources[[6]] is valid.
## ✔ resources[[7]] is valid.
## ✔ resources[[8]] is valid.
## ✔ taxonomic is valid.
## ✔ taxonomic[[1]] is valid.
## ✔ spatial is valid.
## ! spatial cannot be validated (external schema).
## ✔ temporal is valid.
## ✔ temporal$start is valid.
## ✔ temporal$end is valid.
## ✔ Package is consistent with the profile.
##
## ── Check GeoLocator DataPackage Resources
##
## ── Check GeoLocator DataPackage Resources twilights ──
##
## ✔ twilights$tag_id is valid.
## ✔ twilights$twilight is valid.
## ✔ twilights$rise is valid.
## ✔ twilights$label is valid.
## ✔ Table twilights is consistent with the schema.
##
## ── Check GeoLocator DataPackage Resources staps ──
##
## ✔ staps$tag_id is valid.
## ✔ staps$stap_id is valid.
## ✔ staps$start is valid.
## ✔ staps$end is valid.
## ✔ staps$known_lat is valid.
## ✔ staps$known_lon is valid.
## ✔ staps$include is valid.
## ✔ Table staps is consistent with the schema.
##
## ── Check GeoLocator DataPackage Resources paths ──
##
## ✔ paths$tag_id is valid.
## ✔ paths$stap_id is valid.
## ✔ paths$type is valid.
## ✔ paths$lat is valid.
## ✔ paths$lon is valid.
## ✔ paths$ind is valid.
## ✔ paths$j is valid.
## ✔ paths$interp is valid.
## ✔ paths$known is valid.
## ✔ Table paths is consistent with the schema.
##
## ── Check GeoLocator DataPackage Resources edges ──
##
## ✔ edges$tag_id is valid.
## ✔ edges$stap_s is valid.
## ✔ edges$stap_t is valid.
## ✔ edges$lat_s is valid.
## ✔ edges$lon_s is valid.
## ✔ edges$lat_t is valid.
## ✔ edges$lon_t is valid.
## ✔ edges$s is valid.
## ✔ edges$t is valid.
## ✔ edges$j is valid.
## ✔ edges$start is valid.
## ✔ edges$end is valid.
## ✔ edges$n is valid.
## ✔ edges$distance is valid.
## ✔ edges$bearing is valid.
## ✔ edges$gs_u is valid.
## ✔ edges$gs_v is valid.
## ✔ edges$ws_u is valid.
## ✔ edges$ws_v is valid.
## ✔ Table edges is consistent with the schema.
##
## ── Check GeoLocator DataPackage Resources pressurepaths ──
##
## ✔ pressurepaths$tag_id is valid.
## ✔ pressurepaths$datetime is valid.
## ✔ pressurepaths$stap_id is valid.
## ✔ pressurepaths$type is valid.
## ✔ pressurepaths$lat is valid.
## ✔ pressurepaths$lon is valid.
## ✔ pressurepaths$ind is valid.
## ✔ pressurepaths$j is valid.
## ✔ pressurepaths$pressure_tag is valid.
## ✔ pressurepaths$label is valid.
## ✔ pressurepaths$altitude is valid.
## ✔ pressurepaths$surface_pressure is valid.
## ✔ pressurepaths$surface_pressure_norm is valid.
## ✔ pressurepaths$sunset is valid.
## ✔ pressurepaths$sunrise is valid.
## ✔ Table pressurepaths is consistent with the schema.
##
## ── Check GeoLocator DataPackage Resources tags ──
##
## ✔ tags$tag_id is valid.
## ✔ tags$ring_number is valid.
## ✔ tags$scientific_name is valid.
## ✔ tags$manufacturer is valid.
## ✔ tags$model is valid.
## ✔ tags$firmware is valid.
## ✔ tags$weight is valid.
## ✔ tags$attachment_type is valid.
## ✔ tags$readout_method is valid.
## ✔ tags$tag_comments is valid.
## ✔ Table tags is consistent with the schema.
##
## ── Check GeoLocator DataPackage Resources observations ──
##
## ✔ observations$ring_number is valid.
## ✔ observations$tag_id is valid.
## ✔ observations$observation_type is valid.
## ✔ observations$datetime is valid.
## ✔ observations$latitude is valid.
## ✔ observations$longitude is valid.
## ✔ observations$location_name is valid.
## ✔ observations$device_status is valid.
## ✔ observations$observer is valid.
## ✔ observations$catching_method is valid.
## ✔ observations$age_class is valid.
## ✔ observations$sex is valid.
## ✔ observations$condition is valid.
## ✔ observations$mass is valid.
## ✔ observations$wing_length is valid.
## ✔ observations$additional_metric is valid.
## ✔ observations$observation_comments is valid.
## ✔ Table observations is consistent with the schema.
##
## ── Check GeoLocator DataPackage Resources measurements ──
##
## ✔ measurements$tag_id is valid.
## ✔ measurements$sensor is valid.
## ✔ measurements$datetime is valid.
## ✔ measurements$value is valid.
## ✔ measurements$label is valid.
## ✔ Table measurements is consistent with the schema.
## ✔ Package's resources are valid.
##
## ── Check GeoLocator DataPackage Coherence ──
##
## ✔ Package is internally coherent.
##
## ── Check Observations Coherence ──
##
## ✔ observations table is coherent.
##
## ── Overall Package Validation ──
##
## ✔ Package is valid.It is quite normal that you’ll first see some error here. Try to fix them and re-create the package until the package is valid. If you can’t fix them, contact me!
15.5 Upload data to Zenodo
15.5.1 Option 1: Manually
We can write the datapackage to file
write_package(pkg)The content of the folder created can now be uploaded on your Zenodo deposit.
15.5.2 Option 2: Programatically
Alternatively, we can also upload the data to the deposit with
write_package(pkg, directory = pkg$version)
for (f in list.files(pkg$version)) {
zenodo$uploadFile(file.path(pkg$version, f), z)
}15.6 Publish the record
Finally, once you’re happy with everything, you can publish the record on Zenodo